Distance-based sequence logos#
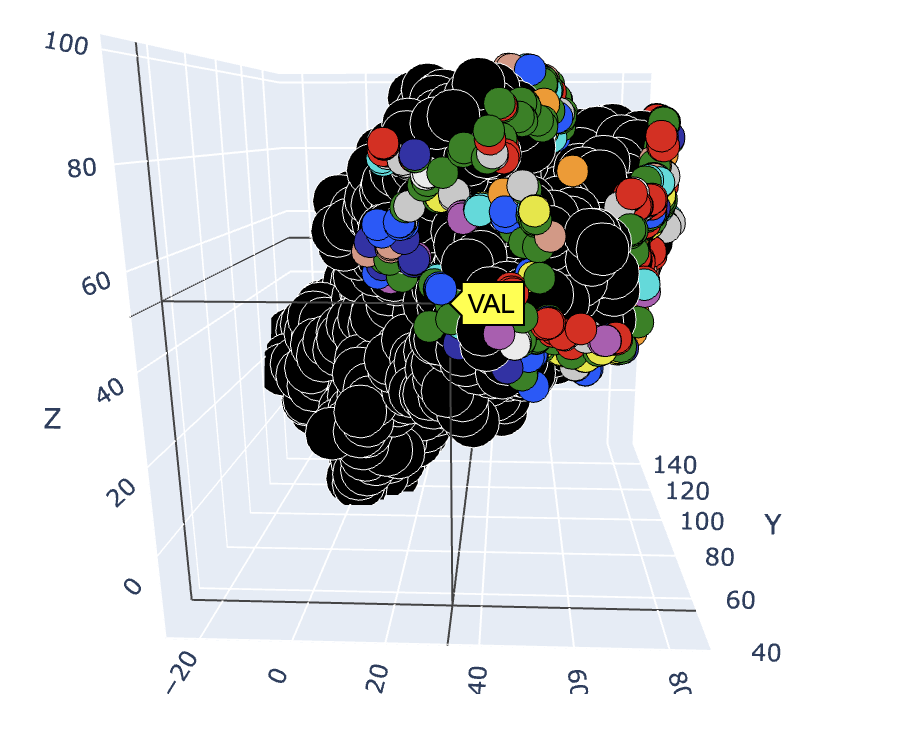
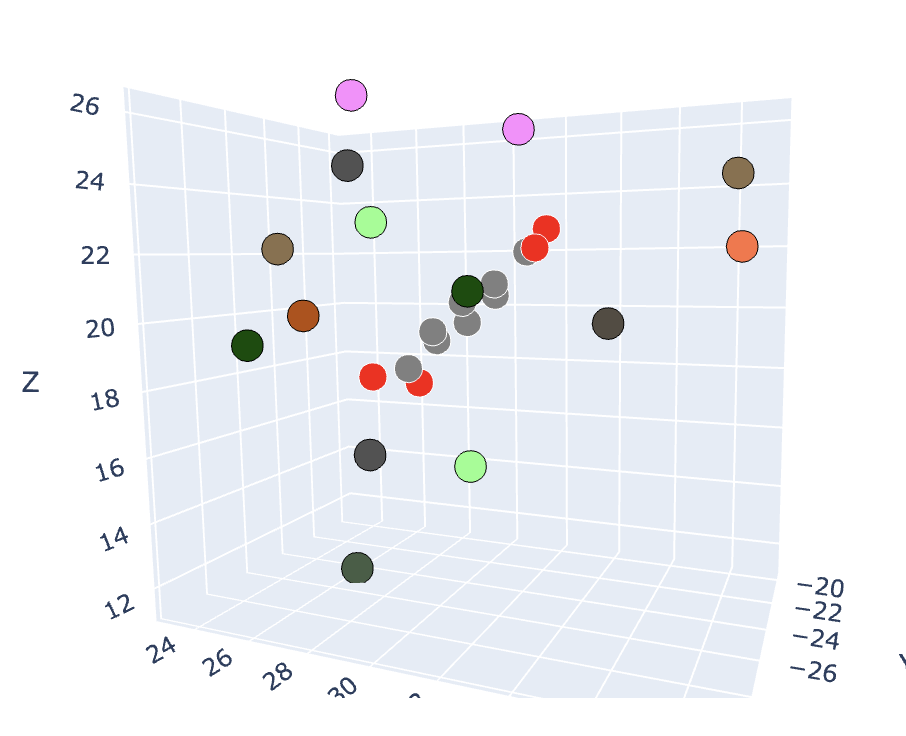
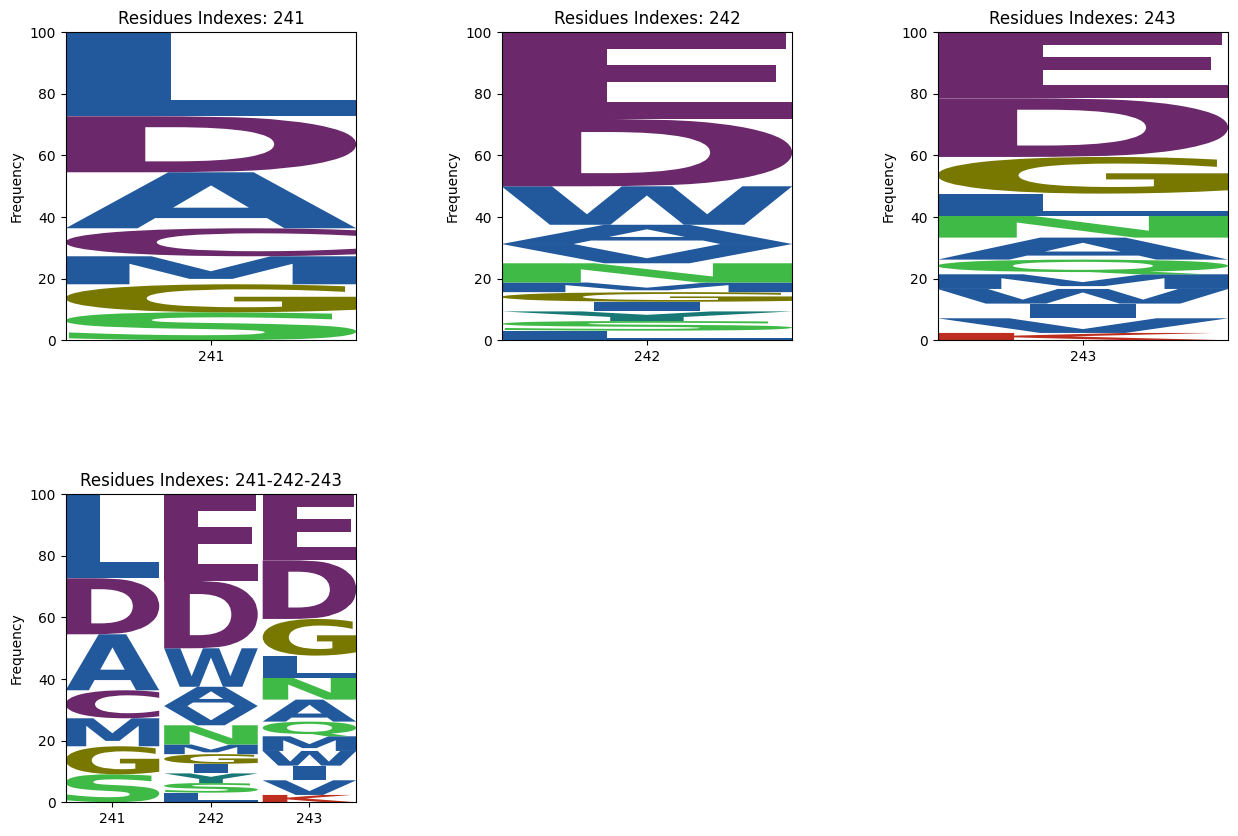
This online tool, hosted on Google Colab, is designed to generate sequence logos based on the distance from a target residue or ligand in a given dataset of pdbs. The distance is calculated using the alpha carbon positions between protein interfaces or all non-hydrogen atoms in the case of ligands. The tool also includes a 3D visualizer that plots the entire target structure and highlights the residues within the specified distance constraint. There are two options for coloring the interacting residues based on Amino colour and Shapely colour.
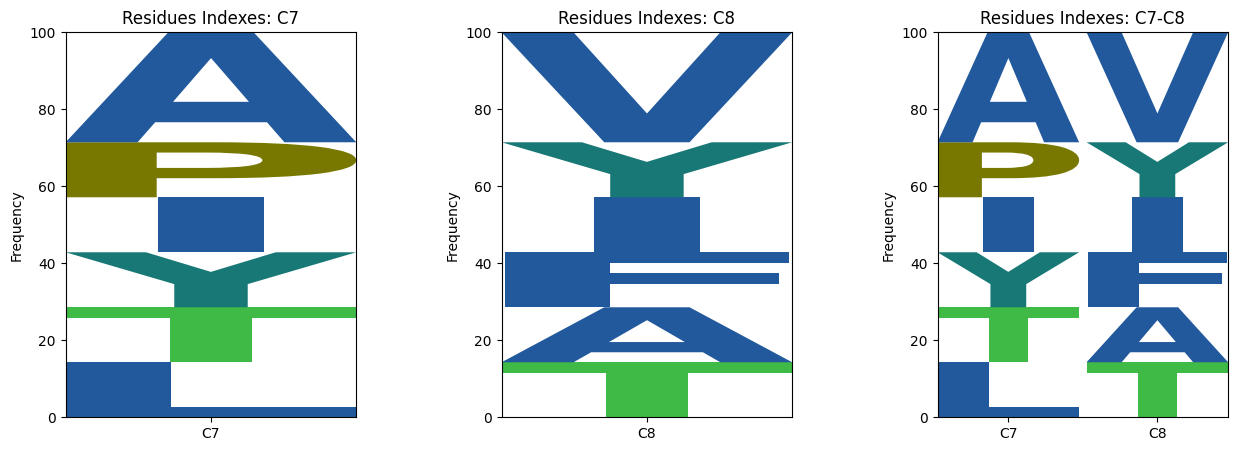
The sequence logos aim to show sequence diversity in binding interactions with various motifs or structures. This tool is able to generate sequence logos and 3d scatter plots from a protein (Figures 1, 3) or a ligand (Figures 2, 4) target.
Figure 1 |
Figure 2 |
|---|---|
|
|
Figure 3 |
Figure 4 |
|---|---|
|
|
Click here to use the interactive Google Colab tool.
Credits:
Daniel:
Discord: theckyy
Kyle:
Discord: kyleweber